Chromosomal Basis of Inheritance | Botany - Answer the following questions (Pure Science Group) | 12th Botany : Chapter 3 : Chromosomal Basis of Inheritance
Chapter: 12th Botany : Chapter 3 : Chromosomal Basis of Inheritance
Answer the following questions (Pure Science Group)
Answer the following questions (Pure Science Group)
Botany : Chromosomal Basis of Inheritance
32. When two different genes came from same parent they tend to remain together.
i) What is the name of this phenomenon?
ii) Draw the cross with suitable example.
iii) Write the observed phenotypic ratio.
Answer: (i) The phenomenon is called linkage.
(ii) William Bateson and Reginald C. Punnet crossed one
homozygous strain of sweet peas having purple
flowers and long pollen grains
with another homozygous strain having red
flowers and round pollen grains.
Results of Test Cross showed that a greater number of F2 plants had purple flowers and
long pollen or red flowers and round pollen. So they concluded that genes for
purple colour and long pollen grain and the genes for red colour and round
pollen grain were found close together in the same homologous pair of
chromosomes. This type of tendency of
genes to stay together during separation of chromosomes is called Linkage.
Alleles in coupling or cis configuration
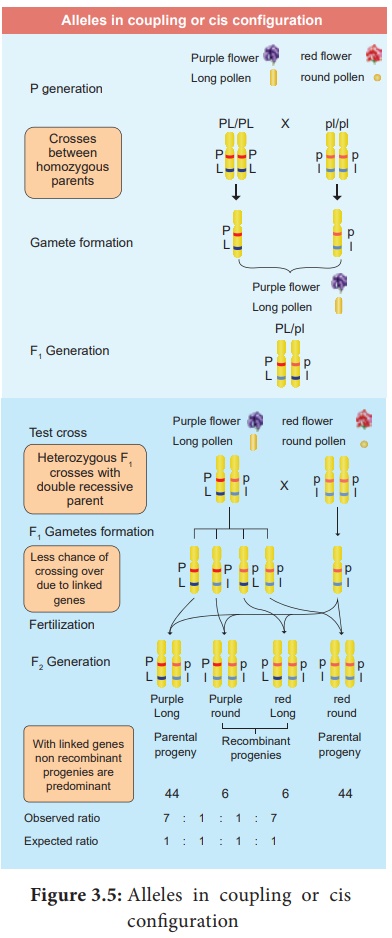
(iii) Phenotypic ratio 7 : 1 : 1 : 7
33. If you cross dominant genotype PV/PV male Drosophila with double recessive female and obtain F1 hybrid. Now you cross F1 male with double recessive female.
i) What type of linkage is seen?
ii) Draw the cross with correct genotype.
iii) What is the possible genotype in F2 generation?
Answer: (i) Complete linkage
(The tendency of genes
to stay together during separation of chromosomes)
(ii)
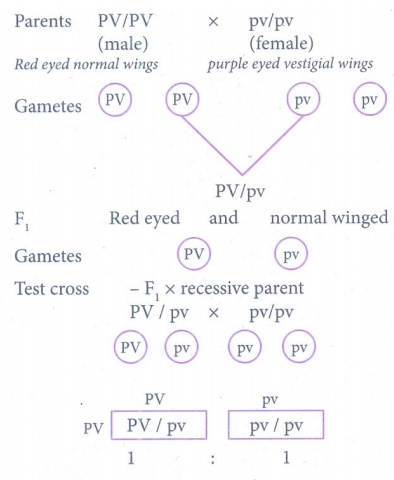
(iii) PV/pv and pv/pv in the ratio 1:1.
34.
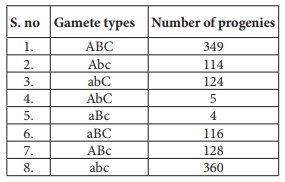
i) What is the name of this test cross?
ii) How will you construct gene mapping from the above given data?
iii) Find out the correct order of genes.
Answer: (i) Three point test cross - gene mapping.
(ii)
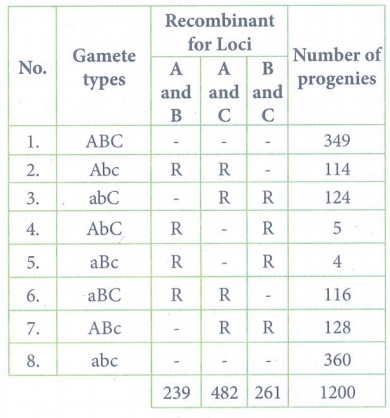
Recombinant frequency
between A and B:
RF = [ Total no. of
recombinants / Total no. of progenies ] x 100
= (239 / 1200) x 100 =
19.9 %
Recombinant frequency
between A and C
RF = (482 / 1200) x 100
= 40.1%
Recombinant frequency
between B and C
RF = (261 / 1200) x
100 = 21.7%
RF is higher between A
and C, so they must be farthest apart. The locus B must lie between them.
Gene mapping:
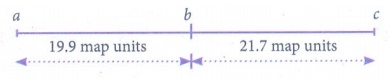
(iii) ABC / abc
35. What is the difference between missense and nonsense mutation?
Answer: The mutation where the codon for one amino acid is changed
into a codon for another amino acid is called Missense or non-synonymous mutations. The mutations where codon for
one amino acid is changed into a termination or stop codon is called Nonsense mutation.
36. 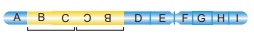
From the above figure identify the type of mutation and explain it.
Answer: The figure shows structural chemical aberration-
Duplication-Reverse tandem type.
Reverse tandem duplication:
The duplicated segment
is located immediately after the normal segment but the gene sequence order
will be reversed.
37. Write the salient features of Sutton and Boveri concept.
Answer: Sutton and Boveri (1903) independently proposed the
chromosome theory of inheritance. Sutton united the knowledge of chromosomal
segregation with Mendelian principles and called it chromosomal theory of
inheritance.
Salient features of the Chromosomal theory of inheritance:
Answer: (i) Somatic
cells of organisms are derived from the zygote by repeated cell division
(mitosis). These consist of two identical sets of chromosomes. One set is
received from female parent (maternal) and the other from male parent
(paternal). These two chromosomes constitute the homologous pair.
(ii) Chromosomes retain their structural uniqueness and
individuality throughout the life cycle of an organism.
(iii) Each chromosome carries specific determiners or Mendelian
factors which are now termed as genes.
(iv) The behaviour of chromosomes during the gamete formation
(meiosis) provides evidence to the fact that genes or factors are located on
chromosomes.
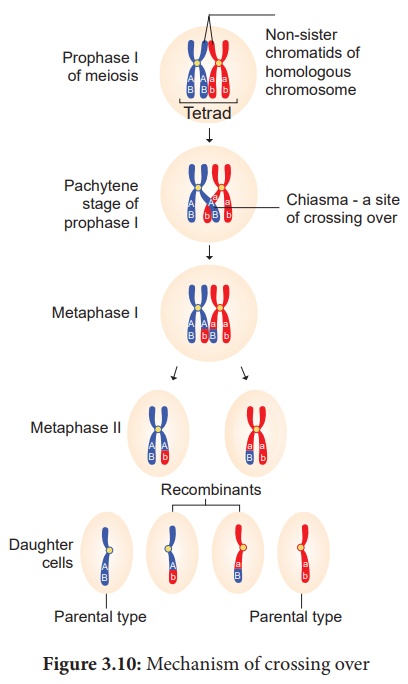
38. Explain the mechanism of crossing over.
Answer: Mechanism of Crossing Over
Crossing over is a
precise process that includes stages like synapsis, tetrad formation, cross
over and terminalization.
(i) Synapsis:
Intimate pairing
between two homologous chromosomes is initiated during zygotene stage of
prophase I of meiosis I. Homologous chromosomes are aligned side by side
resulting in a pair of homologous
chromosomes called bivalents. This pairing phenomenon is called synapsis or syndesis. It is of three types,
1. Procentric synapsis: Pairing starts from middle of the chromosome.
2. Proterminal synapsis: Pairing starts from the telomeres.
3. Random synapsis: Pairing may start from anywhere.
(ii) Tetrad Formation:
Each homologous
chromosome of a bivalent begin to form two identical sister chromatids, which
remain held together by a centromere. At this stage each bivalent has four
chromatids. This stage is called tetrad
stage.
(iii) Cross Over:
After tetrad
formation, crossing over occurs in pachytene stage. The non-sister chromatids
of homologous pair make a contact at one or more points. These points of
contact between non-sister chromatids of homologous chromosomes are called Chiasmata (singular-Chiasma). At
chiasma, cross-shaped or X-shaped structures are formed, where breaking and
rejoining of two chromatids occur. This results in reciprocal exchange of equal
and corresponding segments between them. Synapsis and chiasma formation are
facilitated by a highly organised structure of filaments called Synaptonemal Complex (SC). This
synaptonemal complex formation is absent in some species of male Drosophila.
Hence crossing over does not takes place.
39. Write the steps involved in molecular mechanism of DNA recombination with diagram.
Answer:
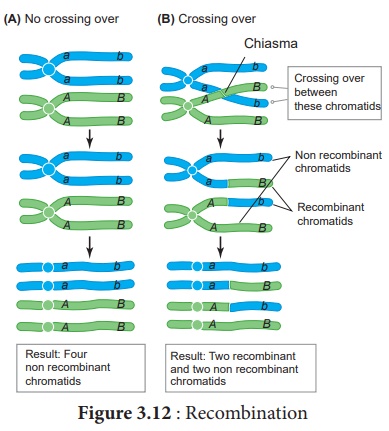
The widely accepted
model of DNA recombination during crossing over is Holliday’s hybrid DNA model.
It was first proposed by Robin Holliday in 1964. It involves several steps. ( Figure
)
(i) Homologous DNA molecules are paired side by side with
their duplicated copies of DNAs.
(ii) One strand of both DNAs cut in one place by the enzyme
endonuclease.
(iii) The cut strands cross and join the homologous strands forming the Holliday
strcture or Holliday junction.
(iv) The Holliday junction migrates away from the original
site, a process called branch migration, as a result heteroduplex region is
formed.
(v) DNA strands may cut along through the vertical (V) line or
horizontal (H) line.
(vi) The vertical cut will result in heteroduplexes with
recombinants.
(vii) The horizontal cut will result in heteroduplex with non
recombinants.
40. How is Nicotiana exhibit self-incompatibility. Explain its mechanism.
Answer: (i) In plants, multiple alleles have been reported in
association with self-sterility or self-incompatibility. Self - sterility means
that the pollen from a plant is unable to germinate on its own stigma and will
not be able to bring about fertilization in the ovules of the same plant.
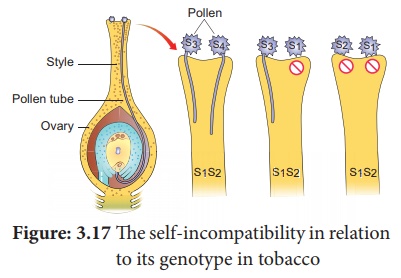
The
self-incompatibility in relation to its genotype in tobacco
(ii) East (1925) observed multiple alleles in Nicotiana which are responsible for
self-incompatibility or self-sterility. The gene for self-incompatibility can
be designated as S, which has allelic series S1, S2, S3,
S4 and S5 .
(iii) The cross-fertilizing tobacco plants were not always
homozygous as S1S1 or S2S2, but all
plants were heterozygous as S1S2, S3S4,
S5S6.
(iv) When crosses were made between different S1S2 plants, the pollen tube did not develop
normally. But effective pollen tube development was observed when crossing was
made with other than S1S2 for example S3S4.
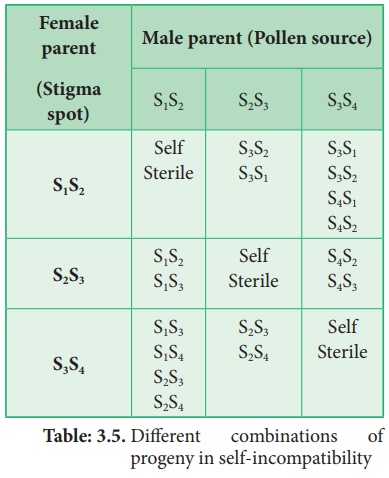
Different combinations
of progeny in self-incompatibility
41. How sex is determined in monoecious plants. write their genes involved in it.
Answer: (i) Zea mays (maize) is an example for monoecious, which means
male and female flowers are present on the same plant.
(ii) There are two types of inflorescence. The terminal
inflorescence which bears staminate florets develops from shoot apical meristem
called tassel. The lateral inflorescence
which develop pistillate florets develops from axillary bud and is called ear or cob.
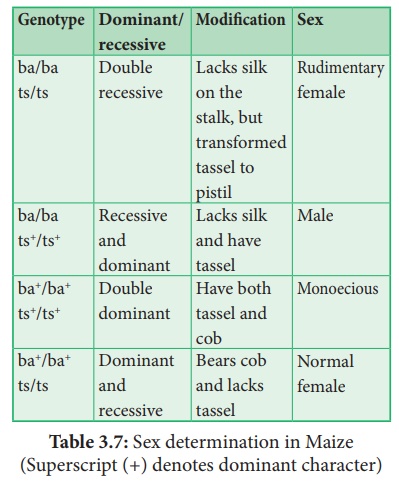
Sex determination in Maize (Superscript (+) denotes
dominant character)
(iii) Unisexuality in maize occurs through the selective abortion
of stamens in ear florets and pistils in tassel florets.
(iv) A substitution of two single gene pairs 'ba' for barren plant and 'ts' for tassel seed makes the difference between monoecious and dioecious (rare) maize plants. The allele for barren plant (ba) when homozygous makes the stalk staminate by eliminating silk and ears. The allele for tassel seed (ts) transforms tassel into a pistillate structure that produce no pollen. Most of these mutations are shown to be defects in gibberellin biosynthesis. Gibberellins play an important role in the suppression of stamens in florets on the ears.
42. What is gene mapping? Write its uses.
Answer: The diagrammatic representation of position of genes and
related distances between the adjacent genes is called genetic mapping. It is directly proportional to the frequency of
recombination between them. It is also called as linkage map. The concept of gene mapping was first developed Alfred H Sturtevant.
Uses of genetic mapping:
(i) It is used to determine gene order, identify the locus of
a gene and calculate the distances between genes.
(ii) They are useful in predicting results of dihybrid and
trihybrid crosses.
(iii) It allows the geneticists to understand the overall
genetic complexity of particular organism.
43. Draw the diagram of different types of aneuploidy.
Answer: It is a condition in which diploid number is altered
either by addition or deletion of one or more chromosomes. Different types of
Aneuploidy is show below. It is classified into two main types - Hyperploidy
and Hypoploidy.
Ploidy
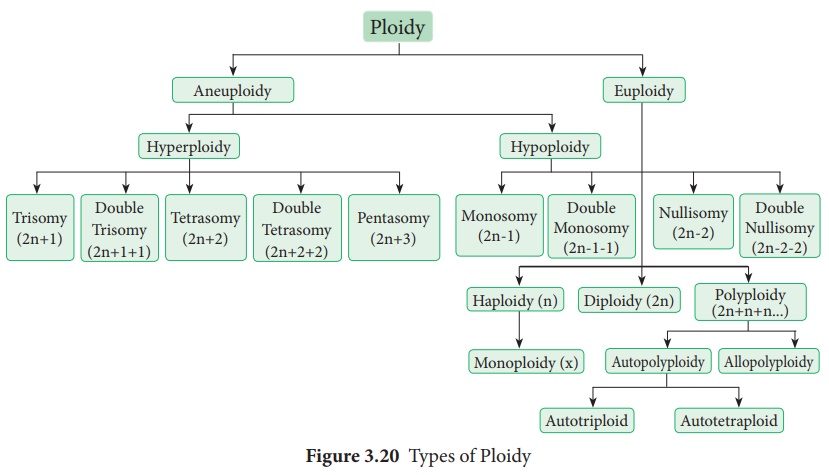
Types of Aneuploidy
44. Mention the name of man-made cereal. How it is formed?
Answer: Triticale is the successful first man made cereal.
Depending on the ploidy level Triticale can be divided into three main groups.
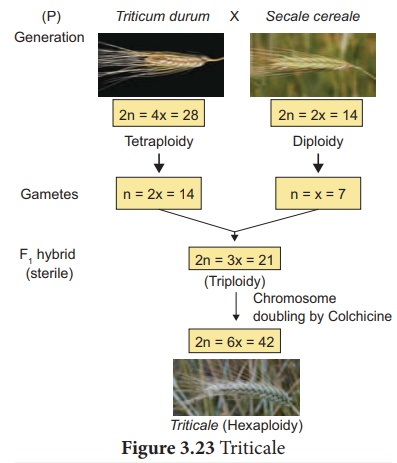
(i) Tetraploidy: Crosses between
diploid wheat and rye.
(ii) Hexaploidy: Crosses between tetraploid wheat Triticum durum (macaroni
wheat) and rye
(iii) Octoploidy: Crosses between hexaploid wheat T. aestivum (bread wheat) and rye.
Hexaploidy: Triticale hybrid plants demonstrate characteristics of
both macaroni wheat and rye. For example, they combine the high-protein content
of wheat with rye’s high content of the amino acid lysine, which is low in
wheat.
Colchicine, an alkaloid when applied in low concentration to the
growing tips of the plants induce polyploidy.
45. What is DNA repair?
Answer: (i) Genomic stability is maintained in all living organisms
which is essential for their survival.
(ii) DNA is unique because it is the only macromolecule where
the repair system exists, which recognises and removes mutations.
(iii) DNA is subjected to various types of damaging reactions
such as spontaneous or environmental agents or natural endogenous threats. Such
damages are corrected by repair enzymes and proteins, immediately after the
damage has taken place.
(iv) DNA repair system plays a major role in maintaining the
genomic / genetic integrity of the organism. DNA repair systems protect the
integrity of genomes from genotoxic stresses.
46. What is replication fork?
Answer: (i) During DNA replication, replication fork is the site
(point of unwinding) of separation of parental DNA strands where new daughter
strands are formed. Multiple replication forks are found in eukaryotes.
(ii) The enzyme helicases are involved in unwinding of DNA by
breaking hydrogen bonds holding the two strands of DNA and replication protein
A (RPA) prevents the separated polynucleotide strand from getting reattached.
47. Write about the energetics of DNA replication.
Answer: The Energetics of
DNA replication - Deoxyribonucleotides such as deoxyadenosine triphosphate
dATP, dGTP, dCTP and dTTP provide energy for the synthesis of DNA. Purpose of
Deoxyribonucelotides (1) acts as a substrate (2) provide energy for polymerisation.
48. What is TATA box?
Answer: In protein synthesis during transcription, Eukaryotic structural gene has 3 features
in promoter
(i) Regulatory elements
(ii) TATA box
(iii) A transcriptional start site
The transcription
start site contains about 25 bp (basepairs) upstream, the sequence is TATAAT
known as TATA or Hogness box which is present in core
promoter.
49. What is alternative splicing?
Answer: It is very useful in regulating gene expression to
overcome the environmental stress in plants.
(i) Alternative splicing is an important mechanism / process
by which multiple mRNA’s and multiple proteins products can be generated from a
single gene. The different proteins generated are called isoforms.
(ii) There are various modes of alternative splicing. When
multiple introns are present in a gene, they are removed separately or as a
unit. In certain cases one or more exons which is present between the introns
are also removed.
Significance of alternative splicing
(i) The proteins transcribed from alternatively spliced mRNA
containing different amino acid sequence lead to the generation of protein
diversity and biological functions.
(ii) Multiple protein isoforms are formed.
(iii) It creates multiple mRNA transcripts from a single gene. A
process of producing related proteins from a single gene thereby the number of
gene products are increased.
(iv) It plays an important role in plant functions such as
stress response and trait selection. The plant adapts or regulates itself to
the changing environment.
50. What is coding strand?
Answer: During protein synthesis, in transcription, the strand of
DNA which is not transcribed is called the coding strand.
51. What are the enzymes involved in DNA replication in eukaryotes?
Answer: (i) The enzyme helicases are involved in unwinding of
DNA by breaking hydrogen bonds holding the two strands of DNA.
(ii) Topoisomerase
is an enzyme which breaks DNAs covalent bonds and removes positive supercoiling
ahead of replication fork.
(iii) DNA replication is initiated by an enzyme DNA polymerase α / primase which
synthesizes short stretch of RNA primers on both leading strand (continuous DNA
strand) and lagging strands (discontinuous DNA strand).
(iv) DNA Pol α (alpha), DNA Pol δ (delta) and DNA Pol ε
(Epsilon) are the 3 enzymes involved in nuclear DNA replication.
(v) DNA Pol α -
Synthesizes short primers of RNA
(vi) DNA Pol δ - Main Replicating enzyme of cell nucleus
(vii)DNA Pol ε - Extend the DNA Strands in replication fork
(viii) DNA ligase joins any nicks in the DNA by forming a
phosphodiester bond between 3’ hydroxyl and 5’ phosphate group.
52. Differentiate coding and non coding strand.
Answer: (i) Coding strand / Non-template strand / Sense
Strand
During protein
synthesis in transcription, the strand of DNA which is not transcribed is
called the Coding Strand.
(ii) Template Strand /
Non-Coding Strand / Antisense Strand
During trascription in
protein synthesis, The strand of DNA which is oriented in 3’ → 5’ direction
that serves as a template for the synthesis of mRNA is called template strand.
53. What are splicesomes?
Answer: (i) RNA splicing is a process which involves the cutting or
removing out of introns and knitting of exons. This process takes place in
spherical particles which is a multiprotein complex called SPLICISOMES. It is
approximately 40 - 60 nm in diameter.
(ii) The spliceosomes have many small nuclear ribonucleic acids
(snRNAs) and small nuclear ribonuclear protein particles (snRNPs) which
identify and helps in the removal of introns.
(iii) A spliceosome removes the introns with an enzyme ribozyme.
Now the mature mRNA comes away from the spliceosomes through the nuclear pore
and is transported out from the nucleus into cytoplasm, and gets attached to
ribosome to carry out translation.
54. What is meant by capping and tailing?
Answer: hnRNA or heterogenous nuclear RNA is the precursor of mRNA
modification at the 5’ end of the primary RNA transcript (hnRNA) with
methylguanosine triphosphate is called capping.
Purpose of capping
(i) Protects RNA from degradation
(ii) Capping plays an important role in removal of first intron
in pre mRNA.
(iii) It regulates the mRNA export from the nucleus into the
cytoplasm.
(iv) It helps in binding of mRNA to theribosome.
Tailing/Polyadenylation
The 3’ end of hnRNA is
cleaved by an endonuclease and a string of adenine nucleotides is added to the
3’ end of hnRNA (pre mRNA) is known as Poly (A) tail - Polyadenylation. This
process is called tailing or polyadenylation.
Purpose of tailing
(i) Translation of RNA transcript is facilitated.
(ii) Helps in the synthesis of polypeptides.
(iii) It enhances the mRNA stability in the cytoplasm.
55. What is RNA editing?
Answer: Chemical modification such as base modification,
nucleotide insertion or deletions and nucleotide replacements of mRNA results
in the alteration of amino acid sequence
of protein that is specified is called RNA editing. This results in the
change in the protein coding sequence of RNA following transcription. The
coding properties of the RNA transcript is changed.
56. Explain the DNA replication in eukaryotes.
Answer: (i) Replication starts at a specific site on a DNA sequence
known as the origin of replication. There are more than one origin of
replication in eukaryotes.
(ii) DNA replication in eukaryotes starts with the assembly of a
prereplication complex (preRC) consisting of 14 different proteins.)
(iii) The origin of replication in yeast is called as ARS
sites (Autonomously Replicating Sequences )
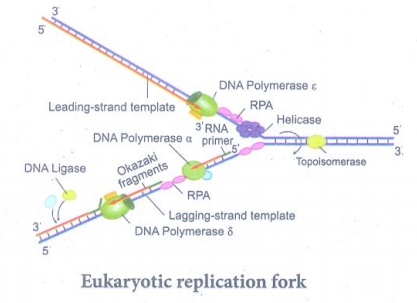
Eukaryotic replication fork
(iv) Replication fork is the site (point of unwinding) of
separation of parental DNA strands where new daughter strands are formed.
Multiple replication forks are found in eukaryotes.
(v) The enzyme helicases
are involved in unwinding of DNA by breaking hydrogen bonds holding the two
strands of DNA and replication protein A (RPA) prevents the separated
polynucleotide strand from getting reattached.
(vi) Topoisomerase is an enzyme which breaks DNAs covalent bonds and removes
positive supercoiling ahead of replication fork.
(vii) DNA replication is initiated by an enzyme DNA polymerase α
/ primase which synthesizes short stretch of RNA primers on both leading strand
(continuous DNA strand) and lagging strands (discontinuous DNA strand).
(viii) Primers are needed because DNA polymerase requires a free
3’ OH to initiate synthesis. DNA polymerase covalently connects the nucleotides
at the growing end of the new DNA strand.
(ix) DNA Pol α (alpha), DNA Pol δ (delta) and DNA Pol ε
(Epsilon) are the 3 enzymes involved in nuclear DNA replication.
(x) DNA Pol α - Synthesizes short primers of RNA.
(xi) DNA Pol δ - Main replicating enzyme of cell nucleus.
(xii) DNA Pol ε - Extend the DNA Strands in replication fork.
(xiii) DNA synthesis takes place in 5’ → 3’ direction and it is
semidiscontinuous. When DNA is synthesized in 5’ → 3’ direction, only in the
free 3’ end (OH end) DNA is elongated.
(xiv) In discontinuous strand the new DNA strand is synthesized
in short pieces called Okazaki fragments. The Okazaki fragments are united by ligase and is also called Lagging
strand where the replication direction is 5’ → 3’ which is opposite to the
direction of fork movement.
(xv) The continuous strand is called Leading strand where the
replication direction is 5’ → 3’ which is same to the direction to that of the
replication fork movement.
(xvi) DNA ligase joins any nicks in the DNA by forming a
phosphodiester bond between 3’ hydroxyl and 5’ phosphate group.
57. With reference to the given diagram correctly match the following pairs.
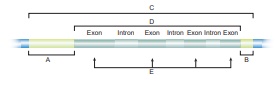
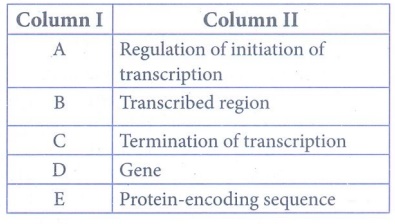
Answer:
Column I - Column
II
A - Gene
B - Transcribed region
C -
Termination of transcription
D - Regulation of initiation of
transcription
E - Protein-encoding sequence
58. What attributes make Arabidopsis a suitable model plant for molecular genetic research?
Answer: Arabidopsis thaliana - Thale cress, Mouse
ear cress
(i) It is a model plant for the study of genetic and molecular
aspects of plant development.
(ii) It belongs to mustard family and it is the first flowering
plant, where its entire genome is sequenced.
(iii) The two regions of the nucleolar organiser ribosomal DNA
which codes for the ribosomal RNA are present at the extremity of chromosomes 2
and 4.
(iv) It is diploid plant having small genome with 2n = 10
chromosomes. Several generations can be produced in one year. So it facilitates
rapid genetic analysis. The genome has low repetitive DNA, over 60% of the
nuclear DNA have protein coding functions.
(v) The plant is small, self fertilizes, annual long-day plant
with short-life cycle. Large numbers of seeds are produced and they are easy to
be grown in laboratory. It is easy to induce mutations. It has many genomic
resources and the transformation can be done easily.
(vi) In 1982, Arabidopsis has successfully completed its life
cycle in microgravity i.e. space. This shows that Human Space Missions with
plant companions may be possible.
59. Describe the molecular mechanism of RNA modification.
Answer: The processing of
pre-mRNA to mature mRNA / Molecular mechanism of RNA modification.
In eukaryotes three
major types of RNA, mRNA, tRNA and rRNA are produced from a precursor RNA
molecule termed as the primary transcript or preRNA. The RNA polymerase II
transcribes the precursor of mRNA, which are also called the heterogenous nuclear RNA or hnRNA which
are processed in the nucleus before they are transported into the cytoplasm.
(i) Capping
Modification at the 5’
end of the primary RNA transcript (hn RNA) with methylguanosine triphosphate is
called capping.
Purpose of capping
(a) Protects RNA from degradation.
(b) Capping plays an important role in removal of first intron
in pre mRNA.
(c) It regulates the mRNA export from the nucleus into the
cytoplasm.
(d) It helps in binding of mRNA to the ribosome.
(ii) Tailing /
Polyadenylation
The 3’ end of hnRNA is
cleaved by an endonuclease and a string of adenine nucleotides is added to the
3’ end of hnRNA (pre mRNA) is known as Poly (A) tail - Polyadenylation. This
process is called tailing or polyadenylation.
Purpose of tailing:
(a) Translation of RNA
transcript is facilitated.
(b) Helps in the synthesis of polypeptides.
(c) It enhances the mRNA stability in the cytoplasm.
The protein coding
regions are not continuous in eukaryotes. Exons
are the coding sequences or expressed sequences contain biological informations
in the matured processed mRNA. Introns are intervening sequences, which are
non-coding sequences (non-amino acid- coding sequences) that should be removed
from a gene before the mRNA product is made. These exons and introns are known
as Split Genes.
(iii) RNA splicing in
plants
(a) RNA splicing is a process which involves the cutting or
removing out of introns and knitting of exons. This process takes place in
spherical particles which is a multiprotein complex called SPLICISOMES. It is
approximately 40 - 60 nm in diameter.
(b) A spliceosome removes the introns with an enzyme ribozyme.
Now the mature mRNA comes away from the spliceosomes through the nuclear pore
and is transported out from the nucleus into cytoplasm, and gets attached to
ribosome to carry out translation.
60. Explain ribosomal translocation in protein synthesis.
Answer: (i) The translation begins with the AUG codon (start codon) of
mRNA.
(ii) Translation occurs on the surface of the macromolecular arena called the
ribosome.
(iii) It is a non-membranous organelle.
(iv) During the process of translation the two sub-units of
ribosomes unite (combine) together and hold mRNA between them.
(v) The protein synthesis begins with the reading of codons of
mRNA.
(vi) The tRNA brings amino acid to the ribosome, a molecular
machine which unites amino acids into a chain according to the information
given by mRNA.
(vii) rRNA plays the structural and catalytic role during translation.
A ribosome has one
binding site for mRNA and two for tRNA. The two binding sites of tRNA are
(i) P-Site - The peptidyl - tRNA binding site is one of the tRNA
binding site. At this site tRNA is held and linked to the growing end of the
polypeptide chain.
(ii) A-Site - The
Aminoacyl - tRNA binding site. This is another tRNA binding site which
holds the incoming amino acids called aminoacyl tRNA. The anticodons of tRNA
pair with the codons of the mRNA in these sites.
Elongation of polypeptide chain
The P and A sites are
near by, so that two tRNA form base pairs with adjacent codon.
The tRNA translates
the genetic code from the nucleic acid sequence to the amino acid sequence i.e
from gene - Polypeptide. When an amino acid is attached to tRNA it is called aminoacylated or charged.
The translation begins
with the AUG codon (start codon) of mRNA. The tRNA which carries first amino
acid methionine attach itself to
P-site of ribosome. The ribosome adds new amino acids to the growing
polypeptides. The second tRNA molecules has anticodons which carries second
amino acid and pairs with the mRNA codon in the A-site of the ribosome. The
first and second amino acids are close enough so that a peptide bond is formed
between them.
The bond between the
first tRNA and methionine now breaks. The first tRNA leaves the ribosome and
the P-site is vacant. The ribosome now moves one codon along the mRNA strand.
The second t-RNA molecule now occupies the P-site. The third t-RNA comes and
fills the A site.
Now a peptide bond is formed
between second and third amino acids. The mRNA then moves through the ribosome
by three bases. This expels deacylated / uncharged tRNA from P-site and moves
peptidyl tRNA into the P-site and empties the A-site. This movement of tRNA
from A-site to P-site is said to be translocation. The translocation requires
thehydrolysis of GTP.
The ribosome (ribozyme - peptidyl transferase) catalyses the
formation of peptide bond by adding amino acid to the growing polypeptide
chain.
The ribosome moves
from codon to codon along the mRNA in the 5’ to 3’ direction. Amino acids are
added one by one, translated into polypeptide as dictated by the mRNA.
Translation is an energy intensive process. A cluster of ribosomes are linked
together by a molecule of mRNA and forming the site of protein synthesis is
called as polysomes or polyribosomes.
61. Describe transposons.
Answer: (i) Transposons
are the DNA sequences which can move from one position to another position in a
genome. This was first reported by an American Geneticist Barbara McClintock as
“mobile controlling element” in Maize.
(ii) Barbara McClintock when studying aleurone of single maize
kernels, noted the unstable inheritance of the mosaic pattern of blue, brown
and red spots due to the differential production of vacuolar anthocyanins.
(iii) In maize plant genome has AC / Ds transposon (AC =
Activator, Ds = Dissociation). The activity of AC element is very distinct in
maize plant. The transposition in somatic cells results in the changes in gene
expression such as variegated pigmentation in maize kernels. Maize genome has
transposable elements which regulated the different colour pattern of kernels.
(iv) McClintock’s findings concluded that Ds and AC genes were
mobile controlling elements. These are called transposable elements.
(v) McClintock gave the first direct experimental evidence
that genomes are not static but are highly plastic entities.
Significance of transposons
(i) They contribute to many visible mutations and mutation
rate in an Organism.
(ii) In evolution, they contribute to genetic diversity.
(iii) In genetic research transposons are valuable tools which
are used as mutagens, as cloning tags, vehicles for inserting foreign DNA into
model organism.
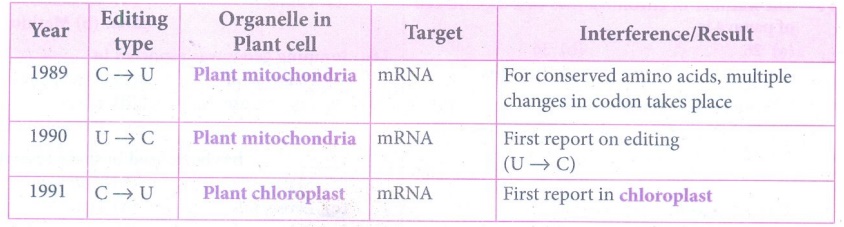
62. Describe RNA editing in plants.
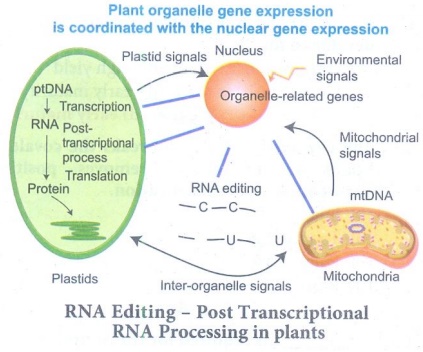
RNA Editing - Post Transcriptional RNA Processing in plants
(i) Chemical modification such as base modification,
nucleotide insertion or deletions and nucleotide replacements of mRNA results
in the alteration of amino acid sequence of protein that is specified is called
RNA editing. This results in the change in the protein coding sequence of RNA
following transcription. The coding properties of the RNA transcript is
changed. The genetic information encoded in the chloroplast genome is altered
by post transcriptional phenomenon which is site - specific (C → U) in
chloroplast of higher plants - RNA editing occurs in plant mitochondria and
chloroplast.
(ii) In plant cells RNA editing by pyrimidine transitions occurs
in mitochondria and plastids (chloroplast). There are two main types of RNA
editing.
1. Substitution editing - Alteration of individual nucleotide bases.
Mitochondria and chloroplast RNA in plants.
2. Insertion / Deletion editing - Nucleotides are
added or deleted from the total number of bases.
Significance of RNA editing
(a) In higher plant chloroplast, it helps to restore the codons
for conserved aminoacids which include initiation and termination codon.
(b) It regulates organellar gene expression in plants.
(c) RNA editing results in the restoration of codons for
phylogenetically conserved amino acid residues.
Related Topics